Publication in Journal of Neural Engineering
https://doi.org/10.1088/1741-2552/abc8d4
Abstract
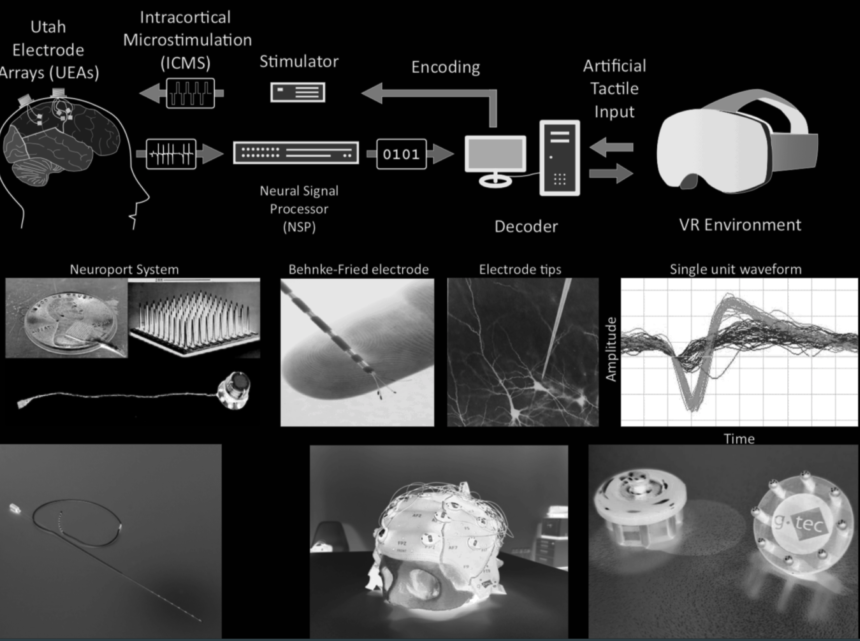
Objective. Advancements in electrode design have resulted in micro-electrode arrays with hundreds of channels for single cell recordings. In the resulting electrophysiological recordings, each implanted electrode can record spike activity (SA) of one or more neurons along with background activity (BA). The aim of this study is to isolate SA of each neural source. This process is called spike sorting or spike classification. Advanced spike sorting algorithms are time consuming because of the human intervention at various stages of the pipeline. Current approaches lack generalization because the values of hyperparameters are not fixed, even for multiple recording sessions of the same subject. In this study, a fully automatic spike sorting algorithm called “SpikeDeep-Classifier” is proposed. The values of hyperparameters remain fixed for all the evaluation data. Approach. The proposed approach is based on our previous study (SpikeDeeptector) and a novel background activity rejector (BAR), which are both supervised learning algorithms and an unsupervised learning algorithm (K-means). SpikeDeeptector and BAR are used to extract meaningful channels and remove BA from the extracted meaningful channels, respectively. The process of clustering becomes straight-forward once the BA is completely removed from the data. Then, K-means with a predefined maximum number of clusters is applied on the remaining data originating from neural sources only. Lastly, a similarity-based criterion and a threshold are used to keep distinct clusters and merge similar looking clusters. The proposed approach is called cluster accept or merge (CAOM) and it has only two hyperparameters (maximum number of clusters and similarity threshold) which are kept fixed for all the evaluation data after tuning. Main Results. We compared the results of our algorithm with ground-truth labels. The algorithm is evaluated on data of human patients and publicly available labeled non-human primates (NHPs) datasets. The average accuracy of BAR on datasets of human patients is 92.3% which is further reduced to 88.03% after (K-means + CAOM). In addition, the average accuracy of BAR on a publicly available labeled dataset of NHPs is 95.40% which reduces to 86.95% after (K-mean + CAOM). Lastly, we compared the performance of the SpikeDeep-Classifier with two human experts, where SpikeDeep-Classifier has produced comparable results. Significance. The results demonstrate that “SpikeDeep-Classifier” possesses the ability to generalize well on a versatile dataset and henceforth provides a generalized well on a versatile dataset and henceforth provides a generalized and fully automated solution to offline spike sorting.